R version 4.3.2 (2023-10-31)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS Sonoma 14.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Asia/Shanghai
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] htmlwidgets_1.6.4 compiler_4.3.2 fastmap_1.1.1 cli_3.6.2
[5] tools_4.3.2 htmltools_0.5.7 rstudioapi_0.15.0 yaml_2.3.8
[9] rmarkdown_2.25 knitr_1.45 jsonlite_1.8.8 xfun_0.41
[13] digest_0.6.34 rlang_1.1.3 evaluate_0.23 在Quarto中嵌入Shiny应用程序
本节不涉及Shiny的教程,请在学习本节前确保已基本掌握Shiny的使用,Shiny官方提供了一个很好的Shiny入门指南,通过它可以对Shiny应用程序的开发有一个全局的了解。

本节主要介绍如何在Quarto文档中嵌入Shiny交互式应用程序,而无需依赖服务器。这是一个Quarto的全新特性,感谢Barret Schloerke等人开发的shinylive包让这一特性得以实现。
Shinylive是一种非服务器依赖的Shiny,它使得Shiny应用程序能够在网页浏览器中直接运行 ,而不需要依赖后台服务器。在 2022 年的Posit大会上,第一次推出了基于 WebAssembly 和 Pyodide 的 Shinylive for Python。随后,在2023年的Posit大会上,首次推出了基于webR的R版本的shinylive。该包的第一个CRAN版本于2023年10月11日首次发布。
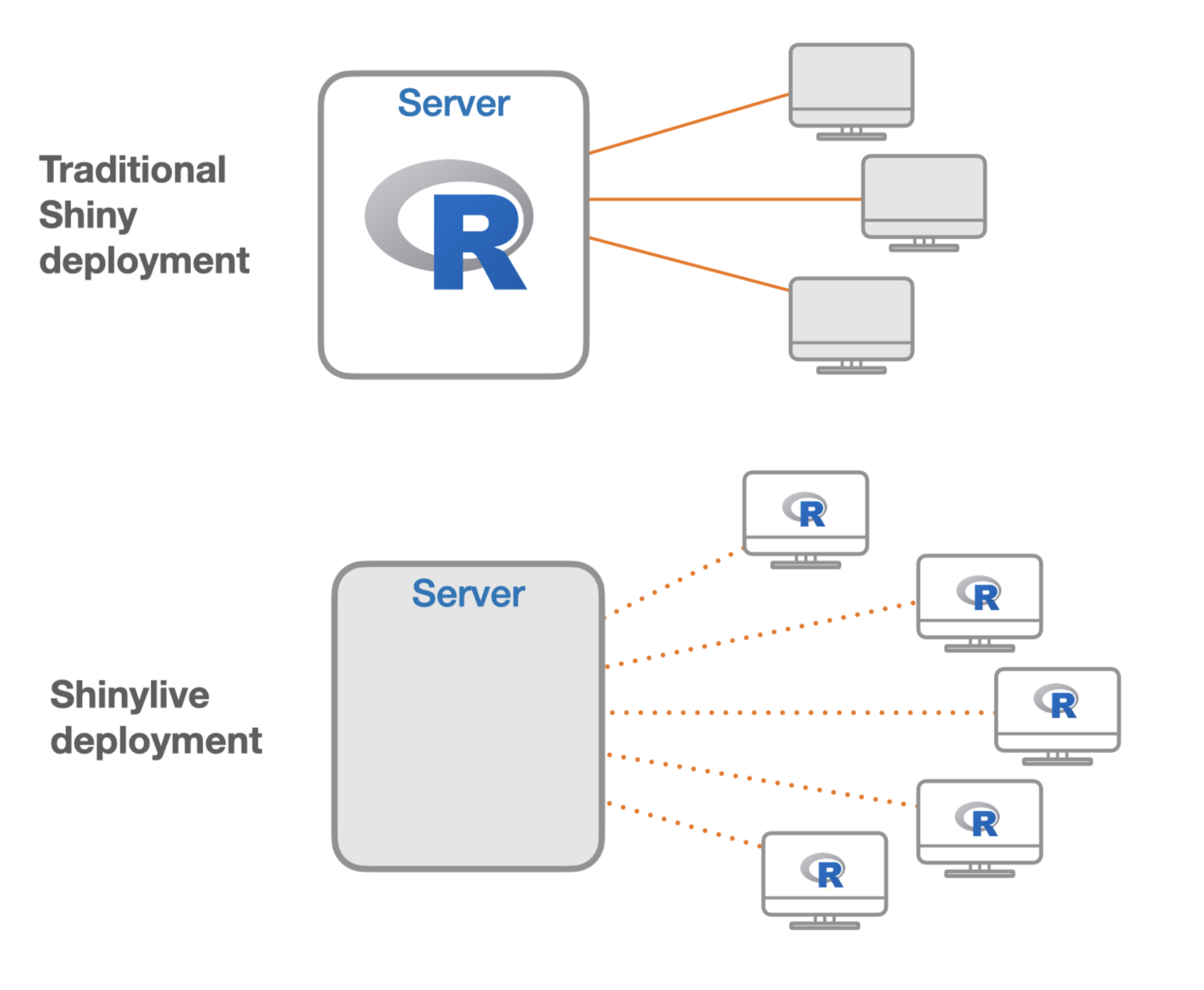
shinylive视频教程《Creating a Serverless R Shiny App using Quarto with R Shinylive》:
Currently, there are three methods (or formats) to use Shinylive applications:
Render a Shiny app into HTML static file using the shinylive package
Host a Shiny app in Fiddle - a built-in web application to run Shiny R and Python applications
Embed Shiny app in Quarto documentation using the quarto-shinylive extension for Quarto (引自:shinylive-r)。
本节我们介绍第三种方法,即在Quarto文档中通过quarto-shinylive extension嵌入Shiny应用程序。
1 安装shinylive包
要实现Shiny应用程序的嵌入,需要依赖shinylive包,可以从CRAN直接安装该包:
install.packages("shinylive")2 建立Quarto Project
创建Quarto项目的方法详见此前的章节。需要注意的是,shinylive 扩展程序必须在 Quarto 项目目录内使用,否则在尝试渲染文档时会报错。错误信息如下:
ERROR:
The shinylive extension must be used in a Quarto project directory
(with a _quarto.yml file).3 安装shinylive扩展程序
为Quarto安装shinylive扩展程序。在Rstudio终端(Terminal)面板中输入:
quarto add quarto-ext/shinylive
4 YAML设置
为了让Quarto能够调用Shiny应用程序,需要在Quarto文档的YAML设置中加上filters-shinylive命令,如下:
title: "Our first r-shinylive Quarto document!"
filters:
- shinylive
---5 编写Shiny程序
我们需要把Shiny程序的代码放到一个特殊的{shinylive-r}代码块内:
```{shinylive-r}
#| standalone: true
library(shiny)
# Define your Shiny UI here
ui <- fluidPage(
# Your UI components go here
)
# Define your Shiny server logic here
server <- function(input, output, session) {
# Your server code goes here
}
# Create and launch the Shiny app
shinyApp(ui, server)
```{shinylive-r}代码块必须包含 #| standalone: true,这表示代码代表了一个完整的独立 Shiny 应用程序。目前,我们需要把完整的Shiny应用程序的代码,包括ui、server等全部包括进一个代码块内。未来,Quarto可能会支持在一个qmd文档内的多个代码块内分别包含ui、server等结构。
6 渲染Quarto文档
编写好所有内容后,我们即可以通过点击“Render”按钮渲染嵌入了Shiny应用程序的Quarto文档了。渲染后,会在我们的Quarto项目根目录下生成一个_extensions文件夹,其结构如下:
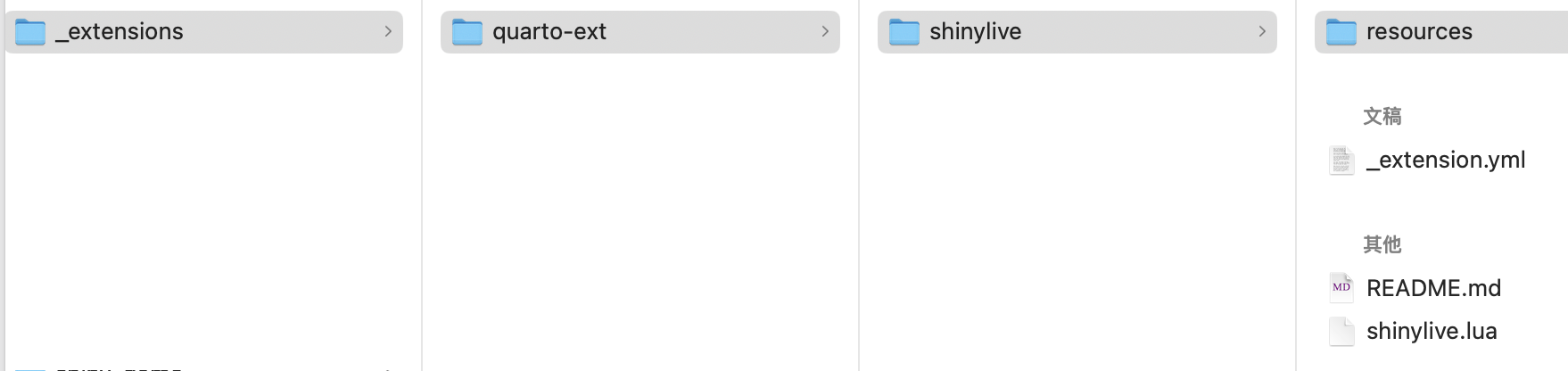
7 发布Quarto文档
一旦制作完成了令人满意的Quarto文档,就可以GitHub Pages和Quarto Pub等通过多种途径发布您的作品了。后面的章节对如何发布GitHub Pages有详细的说明。
更多关于shinylive的信息,参阅:R-shinylive app in Quarto!
8 案例
8.1 案例一
下面是一个来自Shiny官方入门指南中的案例:
#| standalone: true
#| viewerHeight: 600
library(shiny)
# Define UI ----
ui <- fluidPage(
# App title ----
titlePanel("Hello World!"),
# Sidebar layout with input and output definitions ----
sidebarLayout(
# Sidebar panel for inputs ----
sidebarPanel(
# Input: Slider for the number of bins ----
sliderInput(inputId = "bins",
label = "Number of bins:",
min = 5,
max = 50,
value = 30)
),
# Main panel for displaying outputs ----
mainPanel(
# Output: Histogram ----
plotOutput(outputId = "distPlot")
)
)
)
# Define server logic required to draw a histogram ----
server <- function(input, output) {
# Histogram of the Old Faithful Geyser Data ----
# with requested number of bins
# This expression that generates a histogram is wrapped in a call
# to renderPlot to indicate that:
#
# 1. It is "reactive" and therefore should be automatically
# re-executed when inputs (input$bins) change
# 2. Its output type is a plot
output$distPlot <- renderPlot({
x <- faithful$waiting
bins <- seq(min(x), max(x), length.out = input$bins + 1)
hist(x, breaks = bins, col = "#007bc2", border = "orange",
xlab = "Waiting time to next eruption (in mins)",
main = "Histogram of waiting times")
})
}
# Run the app ----
shinyApp(ui = ui, server = server)代码:
```{shinylive-r}
#| standalone: true
#| viewerHeight: 600
library(shiny)
library(bslib)
# Define UI for app that draws a histogram ----
ui <- page_sidebar(
sidebar = sidebar(open = "open",
numericInput("n", "Sample count", 100),
checkboxInput("pause", "Pause", FALSE),
),
plotOutput("plot", width=1100)
)
server <- function(input, output, session) {
data <- reactive({
input$resample
if (!isTRUE(input$pause)) {
invalidateLater(1000)
}
rnorm(input$n)
})
output$plot <- renderPlot({
hist(data(),
breaks = 40,
xlim = c(-2, 2),
ylim = c(0, 1),
lty = "blank",
xlab = "value",
freq = FALSE,
main = ""
)
x <- seq(from = -2, to = 2, length.out = 500)
y <- dnorm(x)
lines(x, y, lwd=1.5)
lwd <- 5
abline(v=0, col="red", lwd=lwd, lty=2)
abline(v=mean(data()), col="blue", lwd=lwd, lty=1)
legend(legend = c("Normal", "Mean", "Sample mean"),
col = c("black", "red", "blue"),
lty = c(1, 2, 1),
lwd = c(1, lwd, lwd),
x = 1,
y = 0.9
)
}, res=140)
}
# Create Shiny app ----
shinyApp(ui = ui, server = server)
```8.2 案例二
#| standalone: true
#| viewerHeight: 500
#| label: fig-shiny-spline
library(ggplot2)
library(htmltools)
ui <- fluidPage(
fluidRow(
column(8,
sliderInput(
"deg_free",
label = "Spline degrees of freedom:",
min = 3L, value = 3L, max = 8L, step = 1L
)
),
imageOutput("spline_contours", height = "400px")
)
)
server <- function(input, output, session) {
# ------------------------------------------------------------------------
# Input data from remote locations on GitHub
pred_path <-
paste(
"https://raw.githubusercontent.com",
"topepo", "shinylive-in-book-test",
"main", "predicted_values.RData",
sep = "/"
)
data_path <-
paste(
"https://raw.githubusercontent.com",
"topepo", "shinylive-in-book-test",
"main", "sim_val.RData",
sep = "/"
)
rdata_file <- tempfile()
download.file(pred_path, destfile = rdata_file)
load(rdata_file)
download.file(data_path, destfile = rdata_file)
load(rdata_file)
# Set some ranges for the plot
rngs <- list(A = c(-3.3, 3.3), B = c(-4.4, 4.4))
output$spline_contours <-
renderImage({
preds <- predicted_values[predicted_values$deg_free == input$deg_free,]
p <-
ggplot(preds, aes(A, B)) +
# Plot the validation set
geom_point(
data = sim_val,
aes(col = class, pch = class),
alpha = 1 / 2,
cex = 3
) +
# Show the class boundary
geom_contour(
aes(z = .pred_one),
breaks = 1 / 2,
linewidth = 3 / 2,
col = "black"
) +
# Formatting
lims(x = rngs$A, y = rngs$B) +
theme_bw() +
theme(legend.position = "top")
file <-
htmltools::capturePlot(
print(p),
tempfile(fileext = ".svg"),
grDevices::svg,
width = 4,
height = 4
)
list(src = file)
},
deleteFile = TRUE)
}
shinyApp(ui = ui, server = server)代码:
```{shinylive-r}
#| standalone: true
#| viewerHeight: 500
#| label: fig-shiny-spline
library(ggplot2)
library(htmltools)
ui <- fluidPage(
fluidRow(
column(8,
sliderInput(
"deg_free",
label = "Spline degrees of freedom:",
min = 3L, value = 3L, max = 8L, step = 1L
)
),
imageOutput("spline_contours", height = "400px")
)
)
server <- function(input, output, session) {
# ------------------------------------------------------------------------
# Input data from remote locations on GitHub
pred_path <-
paste(
"https://raw.githubusercontent.com",
"topepo", "shinylive-in-book-test",
"main", "predicted_values.RData",
sep = "/"
)
data_path <-
paste(
"https://raw.githubusercontent.com",
"topepo", "shinylive-in-book-test",
"main", "sim_val.RData",
sep = "/"
)
rdata_file <- tempfile()
download.file(pred_path, destfile = rdata_file)
load(rdata_file)
download.file(data_path, destfile = rdata_file)
load(rdata_file)
# Set some ranges for the plot
rngs <- list(A = c(-3.3, 3.3), B = c(-4.4, 4.4))
output$spline_contours <-
renderImage({
preds <- predicted_values[predicted_values$deg_free == input$deg_free,]
p <-
ggplot(preds, aes(A, B)) +
# Plot the validation set
geom_point(
data = sim_val,
aes(col = class, pch = class),
alpha = 1 / 2,
cex = 3
) +
# Show the class boundary
geom_contour(
aes(z = .pred_one),
breaks = 1 / 2,
linewidth = 3 / 2,
col = "black"
) +
# Formatting
lims(x = rngs$A, y = rngs$B) +
theme_bw() +
theme(legend.position = "top")
file <-
htmltools::capturePlot(
print(p),
tempfile(fileext = ".svg"),
grDevices::svg,
width = 4,
height = 4
)
list(src = file)
},
deleteFile = TRUE)
}
shinyApp(ui = ui, server = server)
```